This vignette discusses the performance characteristics of
caugi including a comparison to the popular igraph, bnlearn, dagitty, and ggm. We
focus on the practical trade-offs that arise from different core data
structures and design choices.
The headline is: caugi frontloads computation. That is,
caugi spends more effort when constructing a graph and
preparing indexes, so that queries are wicked fast. The high performance
is also due to the Rust backend.
Design choices
Compressed Sparse Row (CSR) representation
The core data structure in caugi is a compressed sparse
row (CSR) representation of the graph. CSR representations store for
each vertex a contiguous slice of neighbor IDs with a pointer (offset)
array that marks the start/end of each slice. This format is memory
efficient for sparse graphs. The caugi graph object also
stores important query information in the object, leading to parent,
child, and neighbor queries being done in
.
This yields a larger memory footprint, but the trade-off is that queries
are extremely fast.
Mutation and lazy building
The caugi graph objects are expensive to build. This is
the performance downside of using caugi. For each time we
make a modification to a caugi graph object, we need to
rebuild the graph completely since the graph object is immutable by
design. This has complexity
,
where
is the vertex set and
is the edge set.
However, the graph object will only be rebuilt when the user either
calls build() directly or queries the graph. Therefore, you
do not need to worry about wasting compute time by iteratively making
changes to a caugi graph object, as the graph rebuilds
lazily when queried. By doing this, caugi graphs
feel mutable, but, in reality, they are not.
By doing it this way, we ensure - that the graph object is always in a consistent state when queried, and - that queries are as fast as possible, while keeping the user experience smooth.
Comparison
Setup
set.seed(42)We are limiting ourselves to comparing graphs up to size
,
as the conversion to bnlearn and dagitty
become prohibitively slow for larger graphs.
generate_graphs <- function(n, p) {
cg <- caugi::generate_graph(n = n, p = p, class = "DAG")
ig <- caugi::as_igraph(cg)
ggmg <- caugi::as_adjacency(cg)
bng <- caugi::as_bnlearn(cg)
dg <- caugi::as_dagitty(cg)
list(cg = cg, ig = ig, ggmg = ggmg, bng = bng, dg = dg)
}Relational queries
We start with parents/children:
graphs <- generate_graphs(1000, p = 0.25) # dense graph
cg <- graphs$cg
ig <- graphs$ig
ggmg <- graphs$ggmg
bng <- graphs$bng
dg <- graphs$dg
test_node_index <- sample(1000, 1)
test_node_name <- paste0("V", test_node_index)
bm_parents_children <- bench::mark(
caugi = {
caugi::parents(cg, test_node_name)
caugi::children(cg, test_node_name)
},
igraph = {
igraph::neighbors(ig, test_node_name, mode = "in")
igraph::neighbors(ig, test_node_name, mode = "out")
},
bnlearn = {
bnlearn::parents(bng, test_node_name)
bnlearn::children(bng, test_node_name)
},
ggm = {
ggm::pa(test_node_name, ggmg)
ggm::ch(test_node_name, ggmg)
},
dagitty = {
dagitty::parents(dg, test_node_name)
dagitty::children(dg, test_node_name)
},
check = FALSE # igraph returns igraph object
)
plot(bm_parents_children)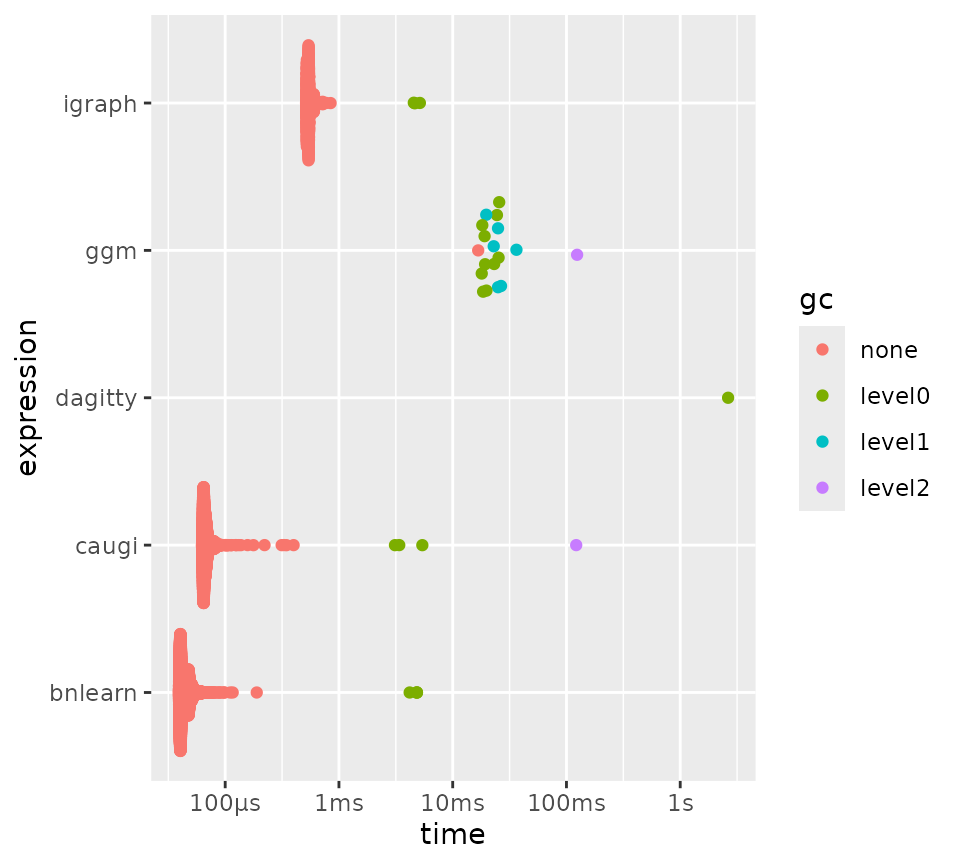
Benchmarking parents/children queries for different packages.
As you can see, bnlearn performs best for this
particular example. In our next experiment, however, we will examine if
this extends to different graph sizes and densities, by parameterizing
our benchmark over n and p. Note that we
adjust p as a function of n to keep the graphs
reasonably sparse.
bm_parents_children_np <-
bench::press(
n = c(10, 100, 500, 1000, 5000, 10000),
p = c(0.5, 0.9),
{
p_mod <- 10 * log10(n) / n * p
graphs <- generate_graphs(n, p = p_mod)
cg <- graphs$cg
ig <- graphs$ig
ggmg <- graphs$ggmg
bng <- graphs$bng
dg <- graphs$dg
test_node_index <- sample(n, 1)
test_node_name <- paste0("V", test_node_index)
bench::mark(
caugi = {
caugi::parents(cg, test_node_name)
caugi::children(cg, test_node_name)
},
igraph = {
igraph::neighbors(ig, test_node_name, mode = "in")
igraph::neighbors(ig, test_node_name, mode = "out")
},
bnlearn = {
bnlearn::parents(bng, test_node_name)
bnlearn::children(bng, test_node_name)
},
ggm = {
ggm::pa(test_node_name, ggmg)
ggm::ch(test_node_name, ggmg)
},
dagitty = {
dagitty::parents(dg, test_node_name)
dagitty::children(dg, test_node_name)
},
check = FALSE # igraph returns igraph object
)
},
.quiet = TRUE
)Next, we plot the benchmark results. As you can see,
bnlearn performs worse as graph size and density increases,
whereas caugi is almost unaffected by these parameters,
outperforming all other packages when graphs get larger and denser.
dagitty and ggm perform worst overall, quickly
becoming infeasible for larger graphs.

Parameterized benchmarking of parents/children queries.
For ancestors and descendants, we see that caugi
outperforms all other packages by a several magnitudes, except for
igraph, which it still beats, but by a smaller margin:
bm_ancestors_descendants <- bench::mark(
caugi = {
caugi::ancestors(cg, "V500")
caugi::descendants(cg, "V500")
},
igraph = {
igraph::subcomponent(ig, "V500", mode = "in")
igraph::subcomponent(ig, "V500", mode = "out")
},
bnlearn = {
bnlearn::ancestors(bng, "V500")
bnlearn::descendants(bng, "V500")
},
dagitty = {
dagitty::ancestors(dg, "V500")
dagitty::descendants(dg, "V500")
},
check = FALSE # dagitty returns V500 as well and igraph returns an igraph
)
plot(bm_ancestors_descendants)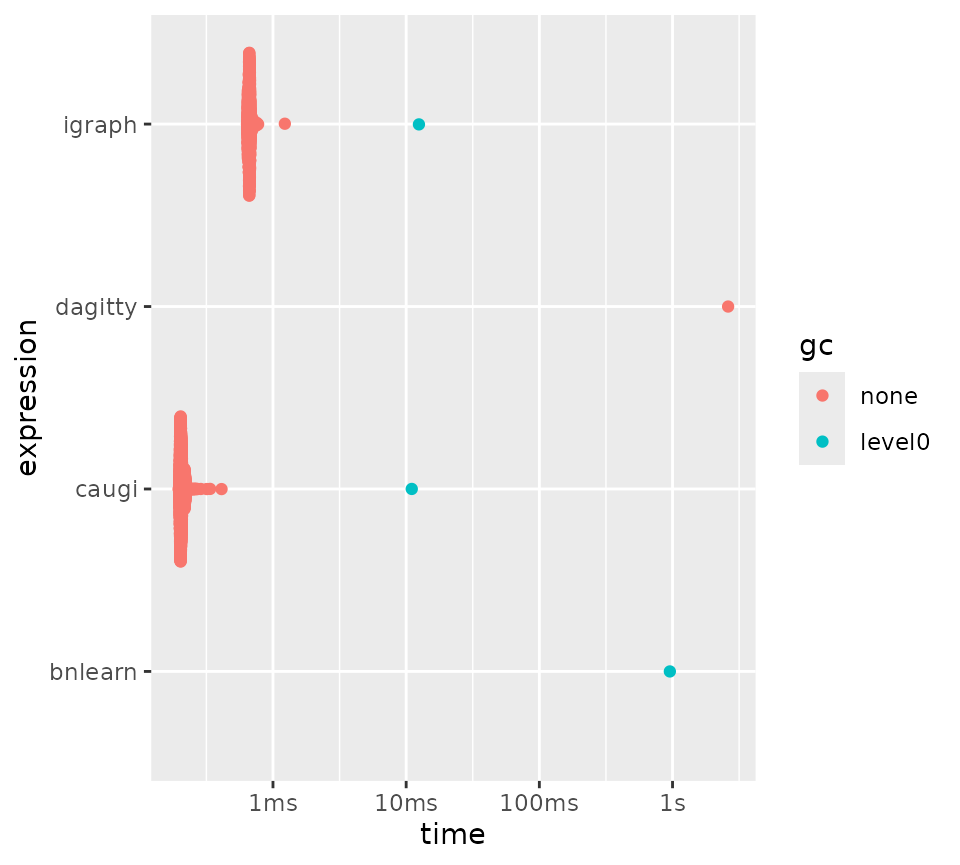
Benchmarking ancestors/descendants queries for different packages.
D-separation
Using the graph from before, we obtain a valid adjustment set and then check for d-separation.
valid_adjustment_set <- caugi::adjustment_set(
cg,
"V500",
"V681",
type = "backdoor"
)
valid_adjustment_set
#> [1] "V7" "V15" "V18" "V24" "V33" "V34" "V68" "V88" "V93"
#> [10] "V121" "V134" "V157" "V168" "V197" "V202" "V206" "V213" "V232"
#> [19] "V262" "V287" "V290" "V291" "V302" "V306" "V314" "V330" "V332"
#> [28] "V342" "V361" "V363" "V368" "V369" "V371" "V385" "V389" "V393"
#> [37] "V409" "V416" "V422" "V425" "V429" "V437" "V444" "V457" "V469"
#> [46] "V476" "V480" "V493" "V504" "V510" "V514" "V522" "V530" "V543"
#> [55] "V555" "V557" "V560" "V564" "V570" "V581" "V588" "V596" "V607"
#> [64] "V620" "V621" "V636" "V641" "V652" "V653" "V660" "V664" "V669"
#> [73] "V673" "V677" "V697" "V701" "V707" "V735" "V743" "V748" "V768"
#> [82] "V785" "V835" "V836" "V844" "V847" "V890" "V893" "V900" "V911"
#> [91] "V912" "V923" "V924" "V927" "V943" "V950" "V964" "V966" "V973"
#> [100] "V974" "V989" "V991" "V996" "V1000"
bm_dsep <- bench::mark(
caugi = caugi::d_separated(cg, "V500", "V681", valid_adjustment_set),
bnlearn = bnlearn::dsep(bng, "V500", "V681", valid_adjustment_set),
dagitty = dagitty::dseparated(dg, "V500", "V681", valid_adjustment_set)
)
plot(bm_dsep)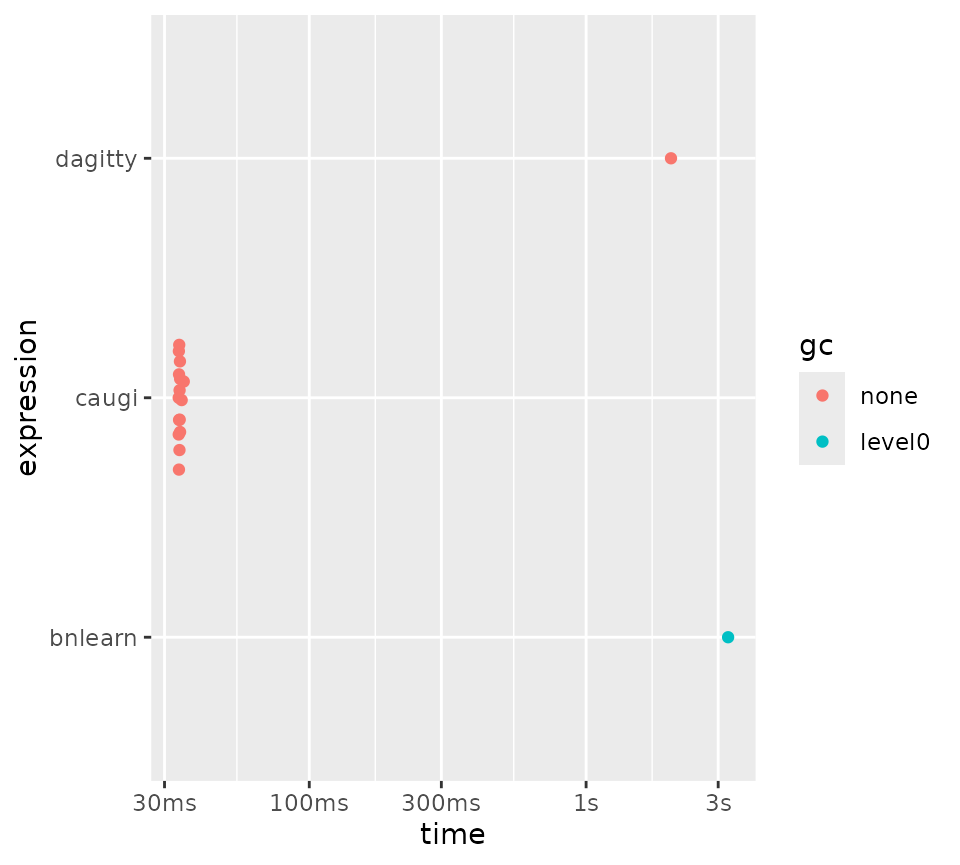
Benchmarks for obtaining a valid adjustment set for d-separation.
Subgraph (building)
Here we see an example of where the frontloading hurts performance.
When we build a subgraph, we have to rebuild the entire
caugi graph object. Here, we see that while
caugi outperforms other packages for queries (except for
parents/children for bnlearn), it is slower for building
the graph objects themselves, which shows in the subgraph benchmark:
subgraph_nodes_index <- sample.int(1000, 500)
subgraph_nodes <- paste0("V", subgraph_nodes_index)
bm_subgraph <- bench::mark(
caugi = {
caugi::subgraph(cg, subgraph_nodes)
},
igraph = {
igraph::subgraph(ig, subgraph_nodes)
},
bnlearn = {
bnlearn::subgraph(bng, subgraph_nodes)
},
check = FALSE
)
plot(bm_subgraph)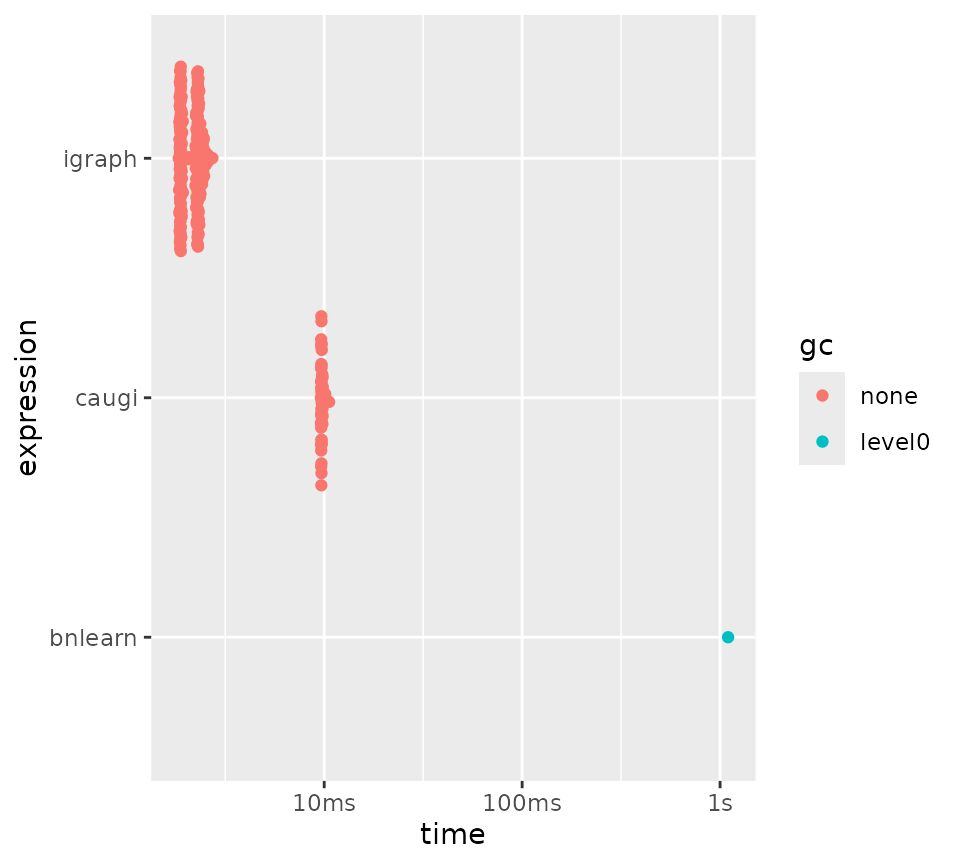
Benchmarking subgraph extraction for different packages.
Session info
sessionInfo()
#> R version 4.5.2 (2025-10-31)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Ubuntu 24.04.3 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] ggplot2_4.0.0
#>
#> loaded via a namespace (and not attached):
#> [1] tidyr_1.3.1 sass_0.4.10 generics_0.1.4
#> [4] digest_0.6.39 magrittr_2.0.4 evaluate_1.0.5
#> [7] grid_4.5.2 RColorBrewer_1.1-3 fastmap_1.2.0
#> [10] jsonlite_2.0.0 graph_1.88.1 bench_1.1.4
#> [13] BiocManager_1.30.27 purrr_1.2.1 dagitty_0.3-4
#> [16] scales_1.4.0 textshaping_1.0.4 jquerylib_0.1.4
#> [19] cli_3.6.5 rlang_1.1.7 ggm_2.5.2
#> [22] bnlearn_5.1 withr_3.0.2 cachem_1.1.0
#> [25] yaml_2.3.12 otel_0.2.0 ggbeeswarm_0.7.2
#> [28] tools_4.5.2 parallel_4.5.2 dplyr_1.1.4
#> [31] profmem_0.7.0 boot_1.3-32 BiocGenerics_0.56.0
#> [34] curl_7.0.0 vctrs_0.7.0 R6_2.6.1
#> [37] stats4_4.5.2 lifecycle_1.0.5 fs_1.6.6
#> [40] V8_8.0.1 htmlwidgets_1.6.4 vipor_0.4.7
#> [43] MASS_7.3-65 ragg_1.5.0 beeswarm_0.4.0
#> [46] pkgconfig_2.0.3 desc_1.4.3 pkgdown_2.2.0
#> [49] pillar_1.11.1 bslib_0.9.0 gtable_0.3.6
#> [52] data.table_1.18.0 glue_1.8.0 Rcpp_1.1.1
#> [55] systemfonts_1.3.1 tidyselect_1.2.1 xfun_0.56
#> [58] tibble_3.3.1 knitr_1.51 farver_2.1.2
#> [61] htmltools_0.5.9 igraph_2.2.1 labeling_0.4.3
#> [64] rmarkdown_2.30 caugi_1.0.0 compiler_4.5.2
#> [67] S7_0.2.1